Research is focussed on understanding the mechanisms by which bacteria sense and respond to environmental stimuli.
In our research we use different strains of Pseudomonas putida and P. aeruginosa as model organisms.
Chemoreceptors and chemotaxis
Identification and analysis of chemoreceptors for TCA cycle intermediates and aromatic pollutants
More recently the laboratory has turned to the study of chemoreceptors and chemotaxis. We have reported the identification of the McpT chemoreceptor for aromatic compounds (Lacal et al., 2011a) and the McpS receptor for Krebs cycle intermediates (Lacal et la., 2010a). In a bioinformatic study we were able to demonstrate that chemoreceptors can be classified according to the size of their ligand binding domain (Lacal et al., 2010b). Efforts are being made to further the functional annotation of chemoreceptors in P. putida and to identify ligands for the mostly uncharacterised chemoreceptors in Pseudomonas.
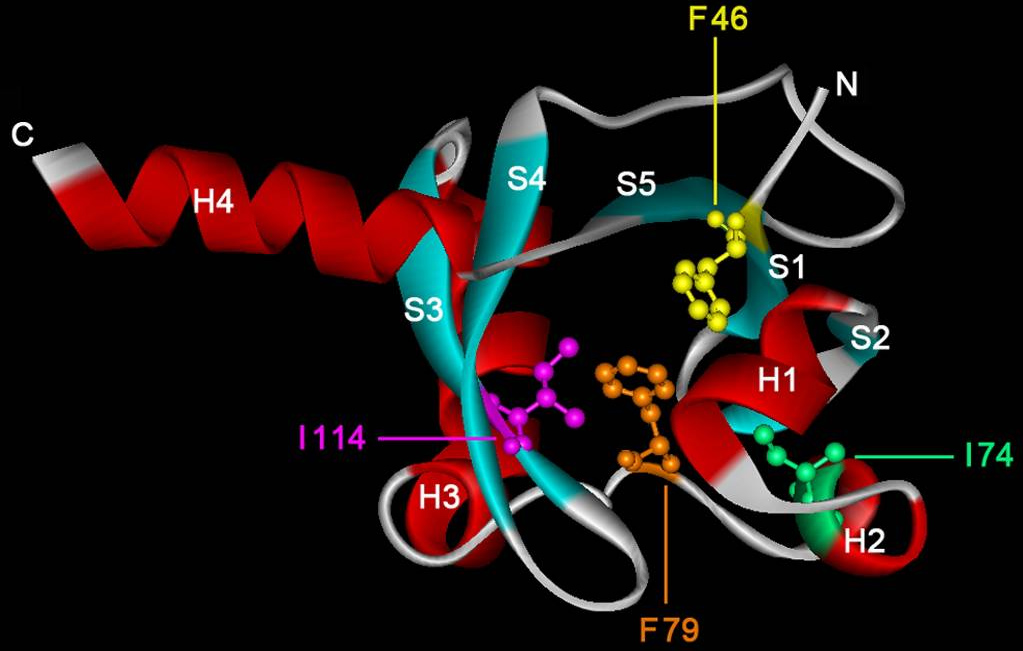
In collaboration with the Laboratory of Crystallographic Studies in Granada we have recently demonstrated that the ligand binding region of the McpS chemoreceptor is composed of two structural modules (Pineda-Molina et al., 2012). We were able to show that chemoattractants bind to each of these modules (see below) and that this results in both cases in a chemotactic response. Current research efforts include the determination of the molecular mechanism of McpS-like receptors and the elucidation of the physiological relevance of chemoreceptors with a bimodular ligand binding region.
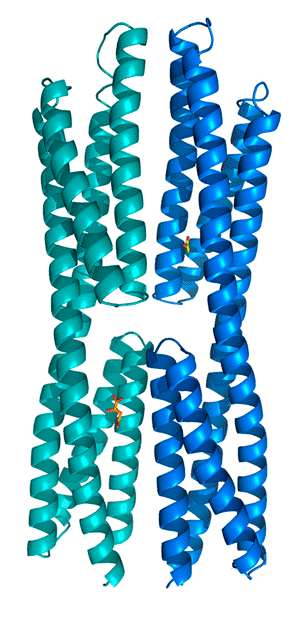
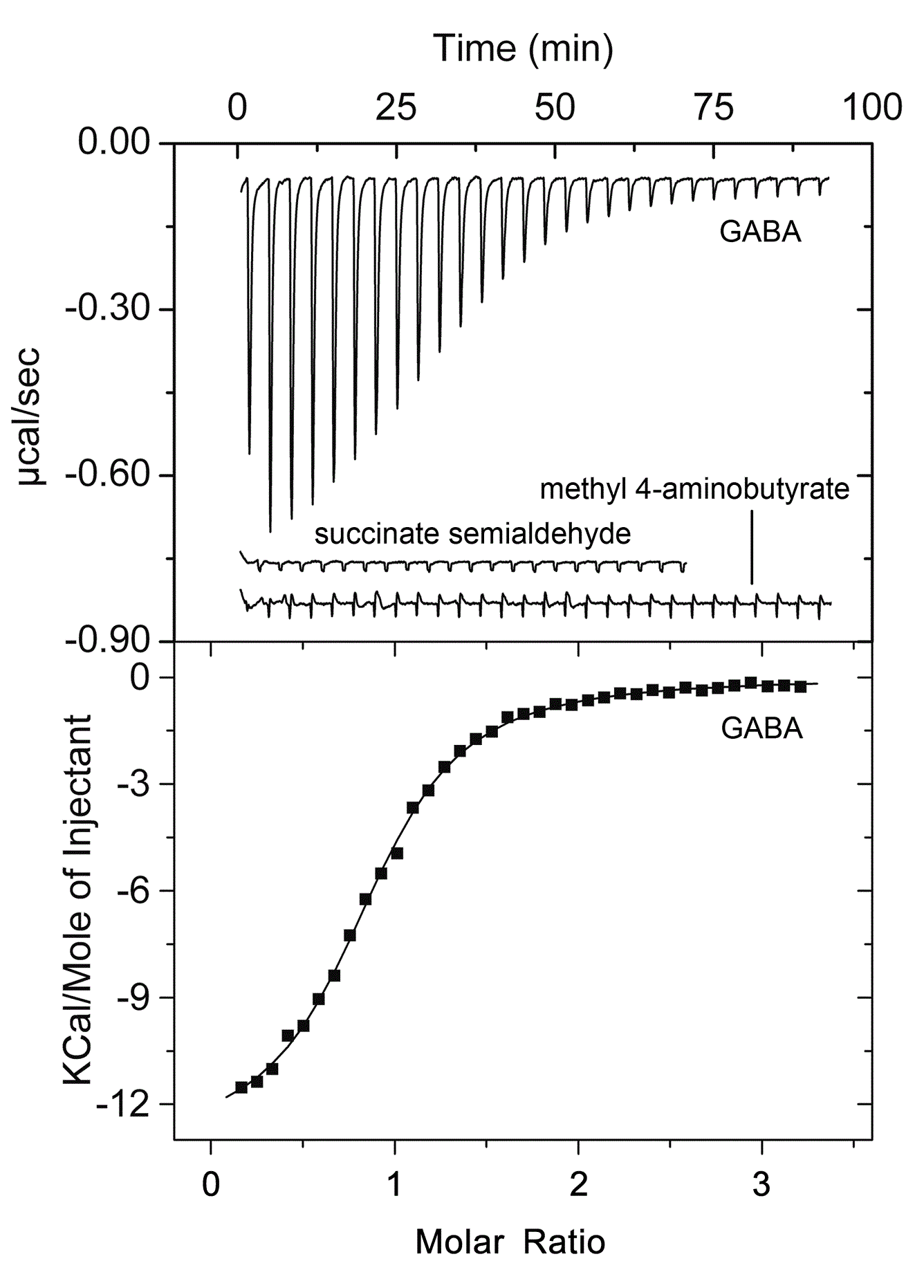
Identification of a chemoreceptor for gamma-aminobutyrate (GABA)
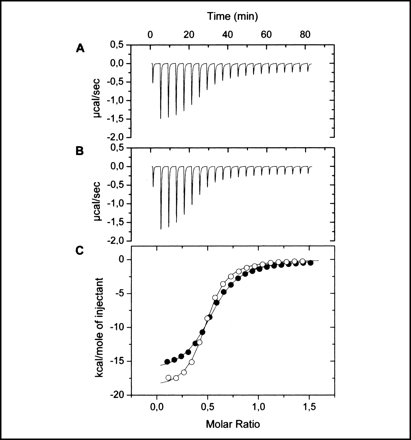
Isothermal titration calorimetry experiments have shown that the ligand binding domain of the PctC chemoreceptor of P. aeruginosa binds GABA with a KD of 1.2 μM. Chemotaxis assays show that the bacterium is attracted to GABA. Since mutation of the pctC gene abolished taxis towards GABA, PctC is the only GABA chemoreceptor of this bacterium. (Rico-Jiménez et al., 2013). We are currently investigating whether other bacteria also possess a GABA-specific chemoreceptor.
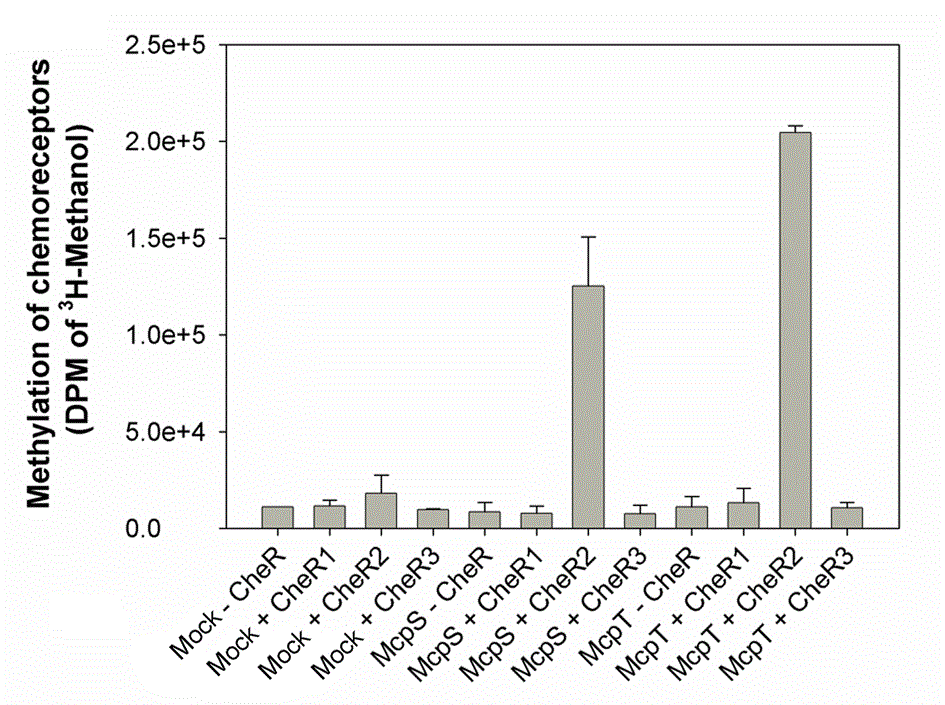
Evaluation of the specificity of the three CheR paralogues in P. putida
The three CheR paralogues of P. putida KT2440 were obtained as recombinant protein and submitted to a series of biophysical analyses and methylation assays. In parallel the three corresponding bacterial mutants were constructed and analyzed (Garcia-fontana et al., 2013). It appeared that the McpS and McpT chemoreceptors are exclusively methylated by CheR2, which is in agreement with chemotaxis studies that showed that mutation in CheR2 abolished taxis. In addition CheR1 played a central role in biofilm formation. No function could yet be attributed to CheR3.
Two component systems
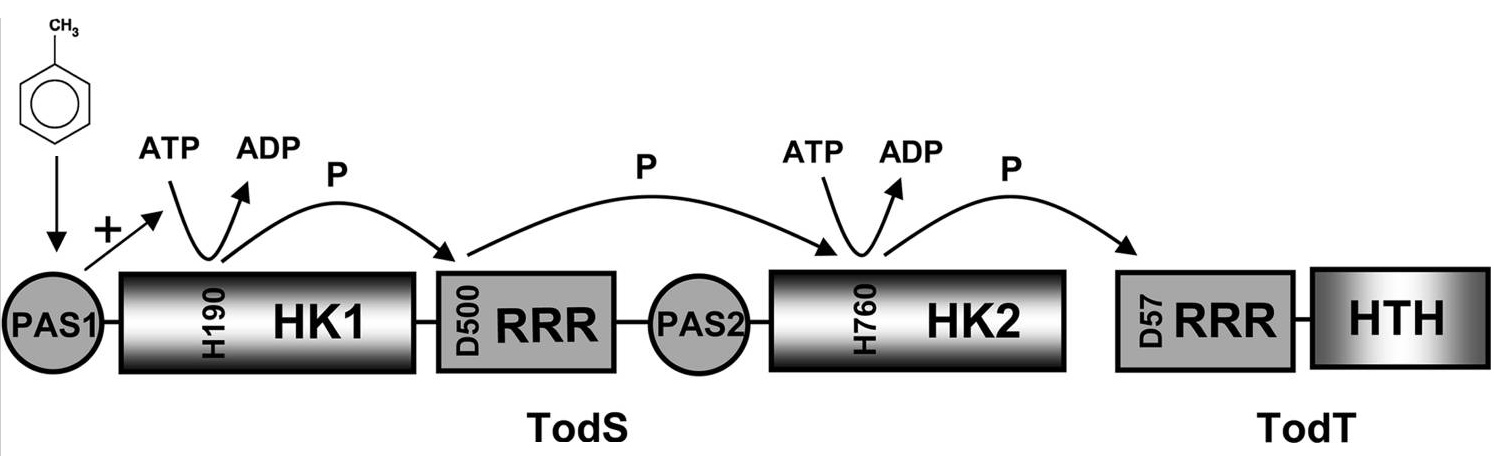
An actual research lines is focussed on the study of members of the TodS/TodT-like family of phosphorelay two-component systems that regulate the expression of degradation pathways for aromatic toxic compounds, like toluene. This line is a continuation of work which has already been reported on the TodS/TodT system (Lacal et al., 2006, Busch et al, 2007, Busch et al., 2009). Current objectives are the identification of the molecular basis underlying the agonistic and antagonistic effect of different effectors on the TodS/TodT system.
One component systems
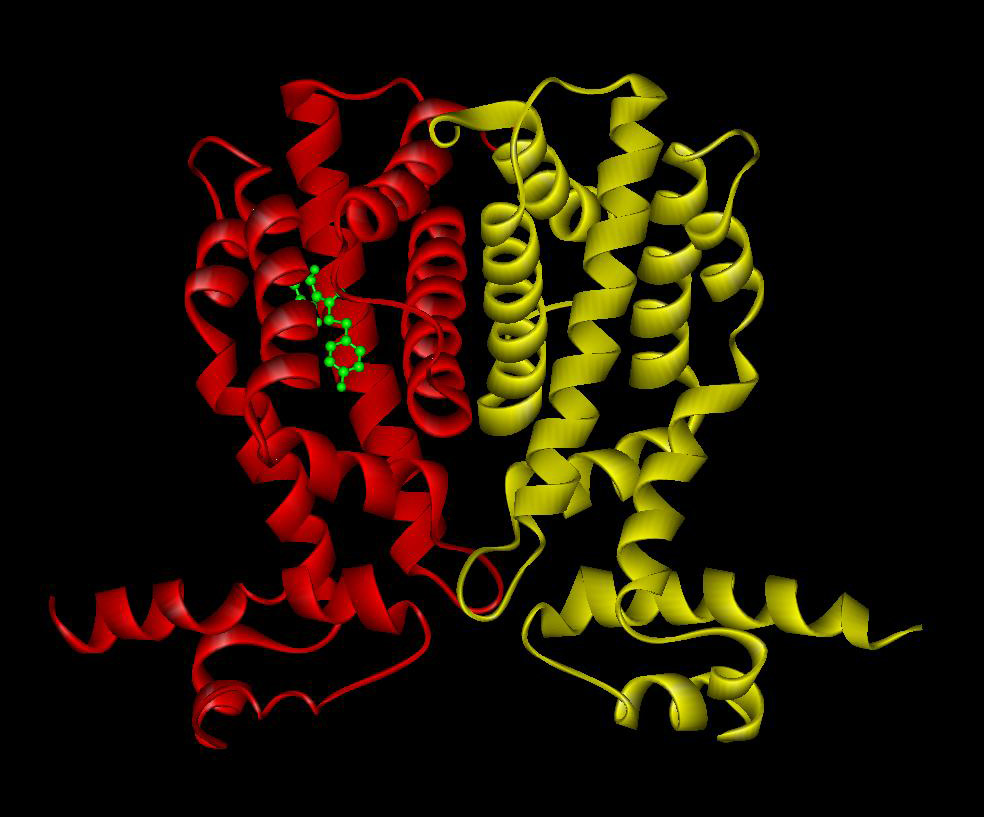
Bacterial signal transduction is primarily mediated by one-component systems, two-component systems and chemoreceptor-based mechanisms. In the past the laboratory has contributed to the study of the TtgR (Terán et al., 2006) and TtgV (Guazzaroni et al., 2007a, Guazzaroni 2007b) one-component systems that mediate the expression of efflux pumps. Work resulted in characterisation of the interaction of both proteins with effector molecules and its DNA operator sites.
Combining in vivo with in vitro experimentation
In our research special emphasis is given to the combination of in vivo with in vitro experimentation. In vivo approaches include the generation and analysis of bacterial mutants, gene expression studies using reporter genes or chemotaxis assays. These approaches are combined with biophysical techniques of biological macromolecules involved in these processes. Techniques so far used include isothermal and differential scanning calorimetry, analytical ultracentrifugation or X-ray diffraction studies to determine protein structures.

