Welcome to the Krell laboratory.
The Krell laboratory is part of the research group “Environmental Microbiology and Biodegradation”. The laboratory is located at the Estación Experimental del Zaidín in Granada (Spain) which is part of the Spanish National Research Council (CSIC). The Krell laboratory participates in the Master “Investigation and Advances in Microbiology” (https://masteres.ugr.es/microbiologia/) at Granada University.
How bacteria recognise stressed plants and move to infection sites
Velando et al. (2025) Genome Biol. 26:260
Glycerol 3-phosphate is an important plant signal molecule. It is synthesised by plants in response to different types of stress, like drought or phosphate starvation. It is also secreted following pathogen attack and accumulates at infection sites. In this study we report the identification of a chemoreceptor, PacP, from the global pathogen Pectobacterium atrosepticum that binds preferentially glycerol 3-phosphate, inducing chemoattraction. Using the combined use of bioinformatic, biochemical and protein structural approaches, we identify a receptor family that binds preferentially glycerol 3-phosphate. Family members were almost exclusively present in plant-associated bacteria, of which many are important pathogens. Taken together, data suggest that this is a mechanism by which bacteria recognise stressed plants and move to infection sites.
Evolution by decreasing complexity
Monteagudo-Cascales et al. (2025) Proc Natl Acad Sci USA 122(5):e2409881122.
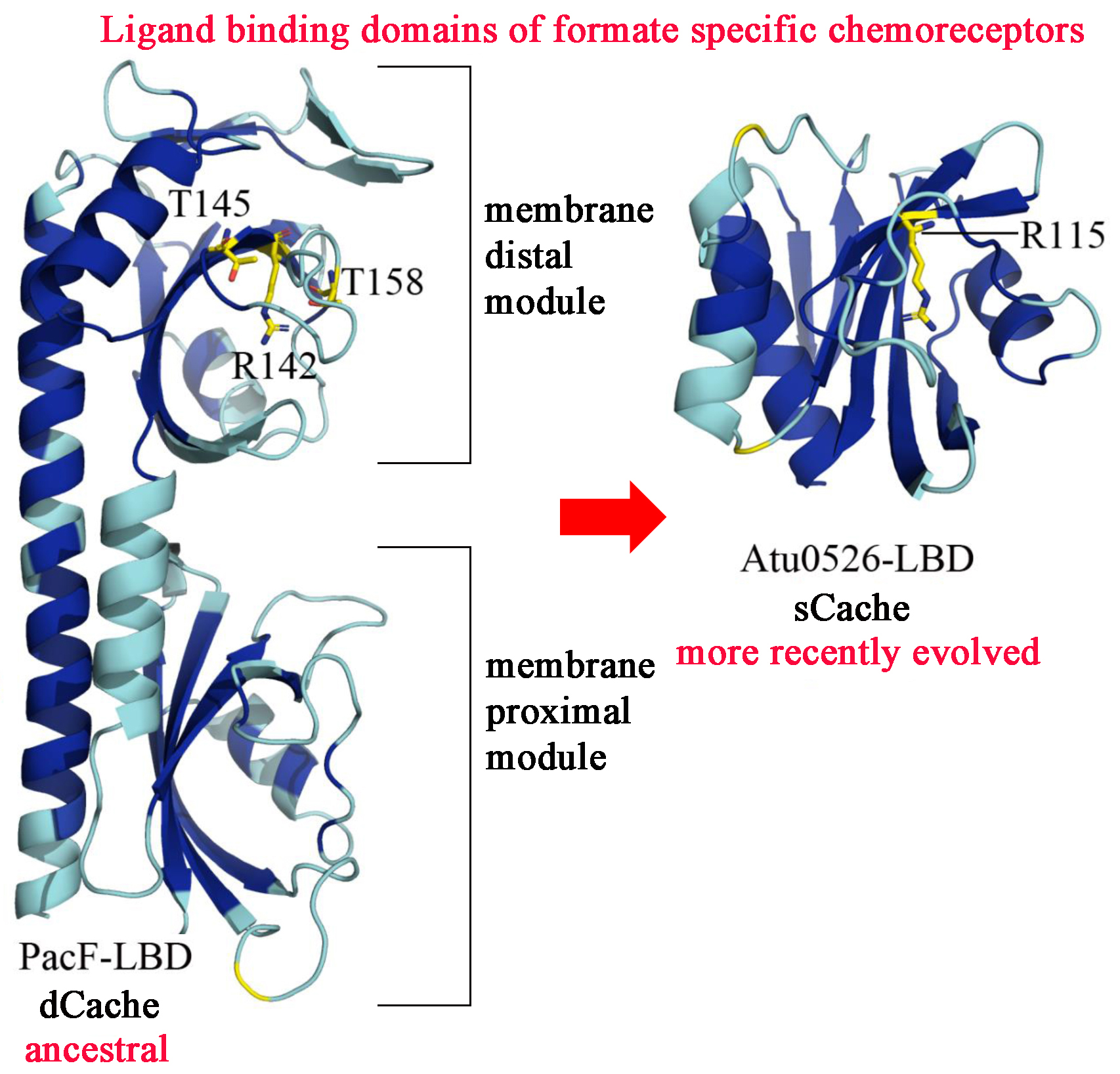
It is generally assumed that evolution is from simple to more complex systems. We show here that this is not always the case. We report on two receptor families in bacteria that bind specifically formate, mediating bacterial chemotaxis. Whereas the ancestral family has a ligand binding domain (LBD) of about 300 amino acids (dCache) that form two structural modules, the more recently evolved receptor family has a LBD that is half in size comprising only a single structural module (sCache). We identify the PacF chemoreceptor from the phytopathogen Pectobacterium atrosepticum that has an ancestral LBD (Figure). The membrane distal module contains the formate binding pocket. The monomodular LBD corresponds to the membrane distal module of PacF. During evolution the membrane proximal module, that is of unknown function, has thus been lost. Both types of LBD bind formate with similar affinity. The characterized family members were highly specific for formate, suggesting that these LBD are well-suited for the development of formate specific chemoreceptors. We report two high resolution X-ray structures of the monomodular LBDs in complex with formate. These structure show that size exclusion, i.e. a binding size that accommodates formate but not any larger molecule, is a mechanism that guarantees ligand specificity. Future studies will show whether this is an isolated case or whether there are similar events that have occurred in the evolution of other receptor families.
Purines are important bacterial signal molecules
Monteagudo-Cascales et al. (2024) Nature Communications 15, 5867
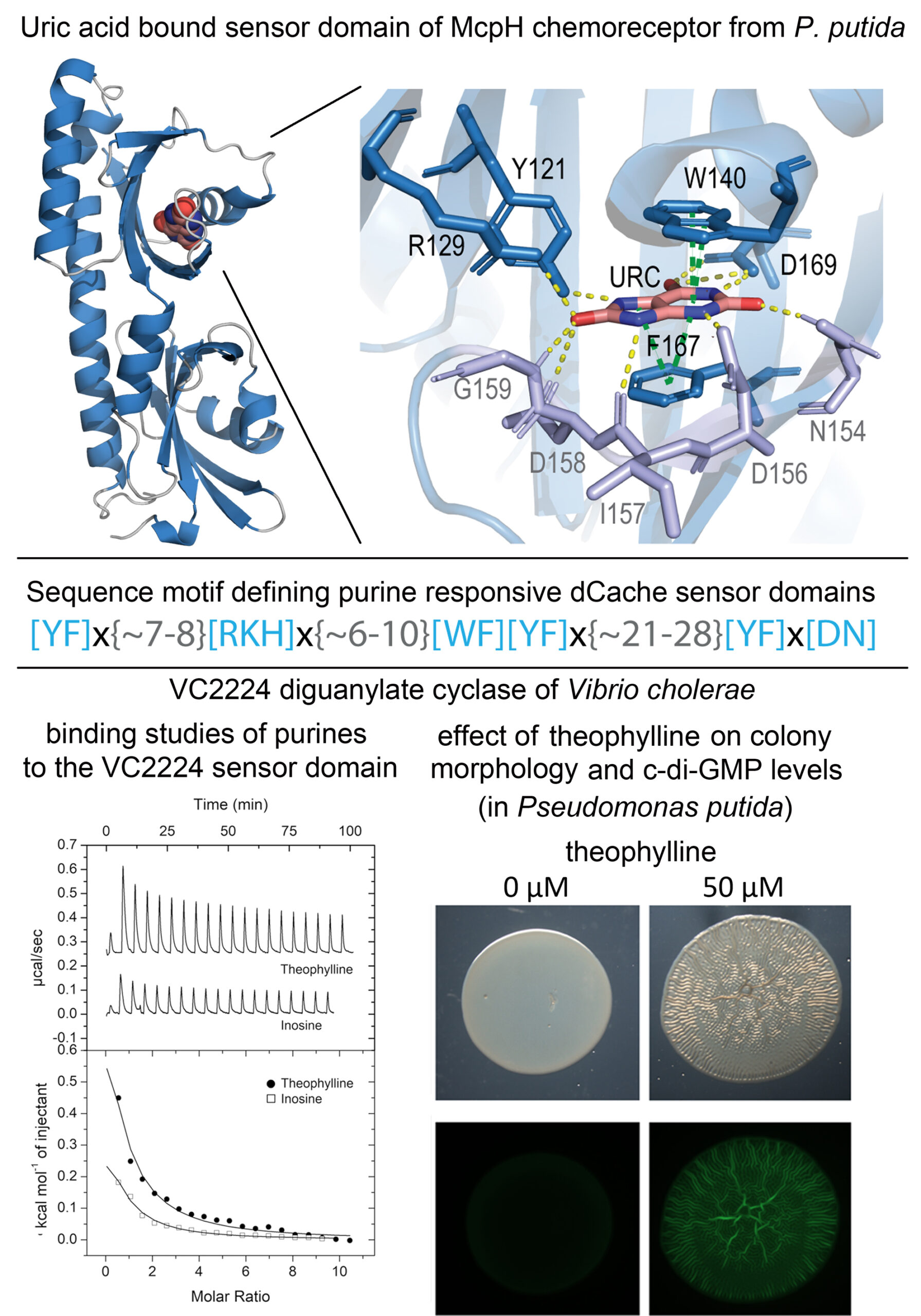
In humans, purines are important signal molecules that control a variety of cellular processes. Very scarce information is available on the signaling role of purins in bacteria. We report the 3D structure of the sensor domain of the purine specific chemoreceptor McpH in complex with bound uric acid and define a sequence motif of amino acids involved in ligand binding. Searches in sequence databases resulted in the retrieval of 6,300 proteins that match this motif constituting the family of purine specific sensor domains (dCache_1PU). Ligand binding studies with selected family members confirm the precision of this prediction. Family members show a wide phylogenetic distribution including important human and plant pathogens. Next to chemoreceptors, dCache_1PU domains were identified in sensor histidine kinases, diguanylate cyclases and phosphodiesterases as well as sin Ser/Thr phosphatases, indicating that purines are important signal molecules that control in bacteria processes like gene expression, metabolism, second messenger levels or chemotaxis. Using the dCache_1PU containing VC2224 diguanylate cyclase, we show that the purine ligands identified induce a wrinkly colony morphology that was caused by increases in c-di-GMP levels. The dCache_1PU adds to the amino acid (Gumerov et al. (2022) PNAS 119:e2110415119) and biogenic amine (Cerna-Vargas et al. (2023) PNAS 120:e2305837120) specific families that we have identified before.
Many bacterial receptors contain sensor domains that bind biogenic amines
Cerna-Vargas et al. (2023) Proc Natl Acad Sci USA 120:e2305837120
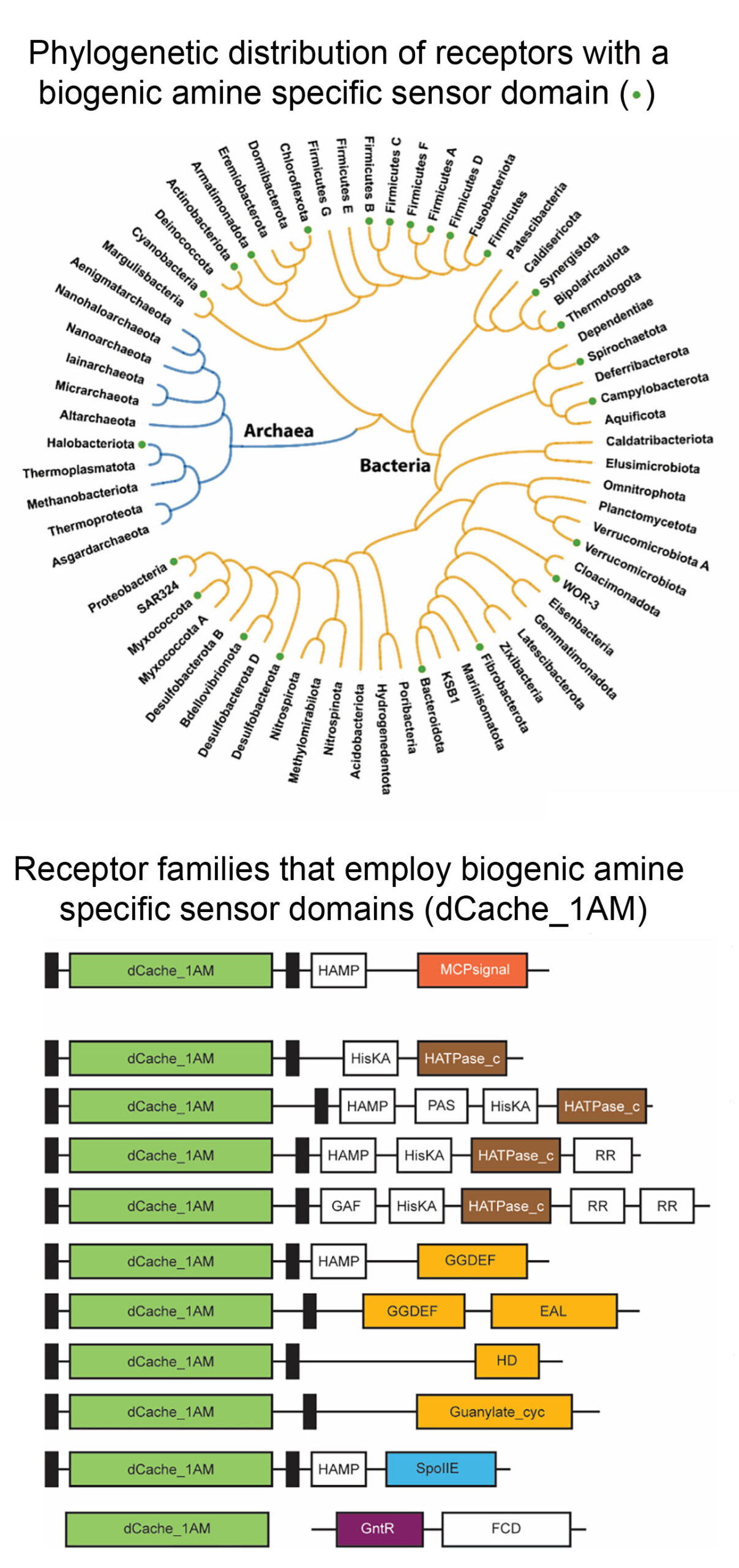
Following an analogous approach that we have used to identify amino acid specific sensor domains (Gumerov et al. (2022) Proc. Natl. Acad. Sci. USA 119:e2110415119), we identify a binding pocket sequence motif that was present in 3D structures of sensor domains crystallised in complex with quaternary amines. Screening of sequence databases for the presence of this motif resulted in the identification of 13.000 different receptors, that formed the basis for the definition of the amine specific subfamily of dCache sensor domains (dCache_1AM). The individual sensor domain of representative receptors was generated as individual purified protein, and their analysis by thermal shift ligand screening and microcalorimetric titrations indicated that the predictions were highly precise. dCache_1AM sensor domains show a wide phylogenetic distribution in bacteria and were also detected in archaea (Fig. below). These domains were identified in a variety of receptors such as chemoreceptors, sensor histidine kinases or diguanylate/guanylate cyclases and phosphohydrolases that differ in function and mediate chemotaxis, regulate gene expression or control second messenger homeostasis, indicating the amines are central bacterial signal molecules. This article has been commented by Dlakić (2023) (Proc. Natl. Acad. Sci. USA 120:e2316830120).
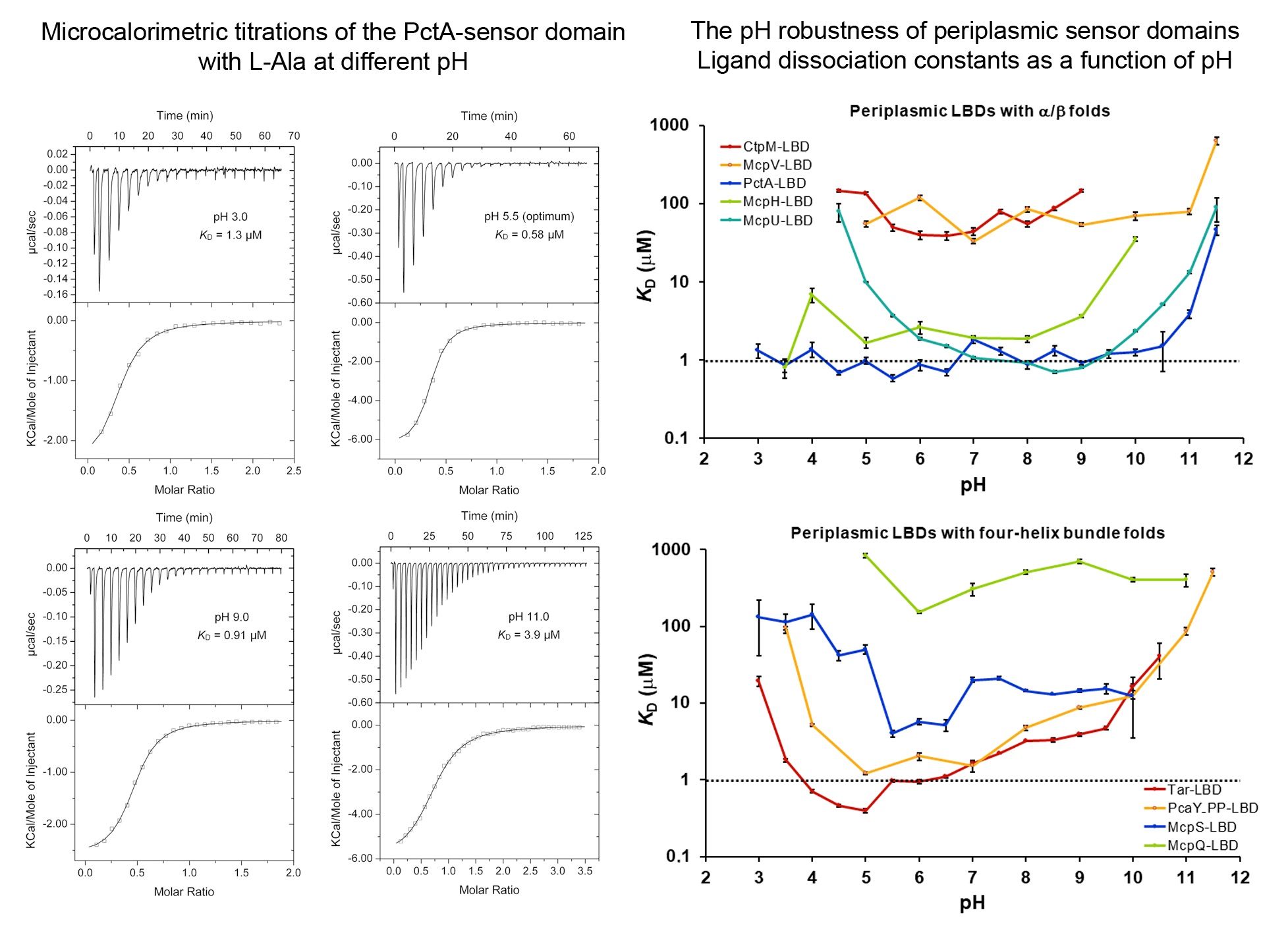
The stunning pH robustness of extracytosolic bacterial sensor domains
Signal transduction is typically initiated by the recognition of signal molecules at receptor sensor domains. Hundreds of different types of sensor domains have evolved. In this article we have investigated the capacity of members of the four major superfamilies of extracytosolic sensor domains to recognize their signals at different pH. We have furthermore analysed the pH dependence of signal recognition of periplasmic solute binding proteins as well as cytosolic sensor domains. We show that extracytosolic sensor domains possess a stunning pH robustness, which is exemplified by the sensor domains of the Tar and PctA chemoreceptors that recognized their cognate signals over 7.5 and 8.5 pH units, respectively. A similar pH robustness was observed for periplasmic solute binding proteins, whereas cytosolic sensor domains bound their ligands over a much narrower pH range. Data thus suggest that many receptors maintain their sensing capacities over a broad range permitting in turn signal integration under harsh environmental conditions. These data also permit the construction of robust biosensors.
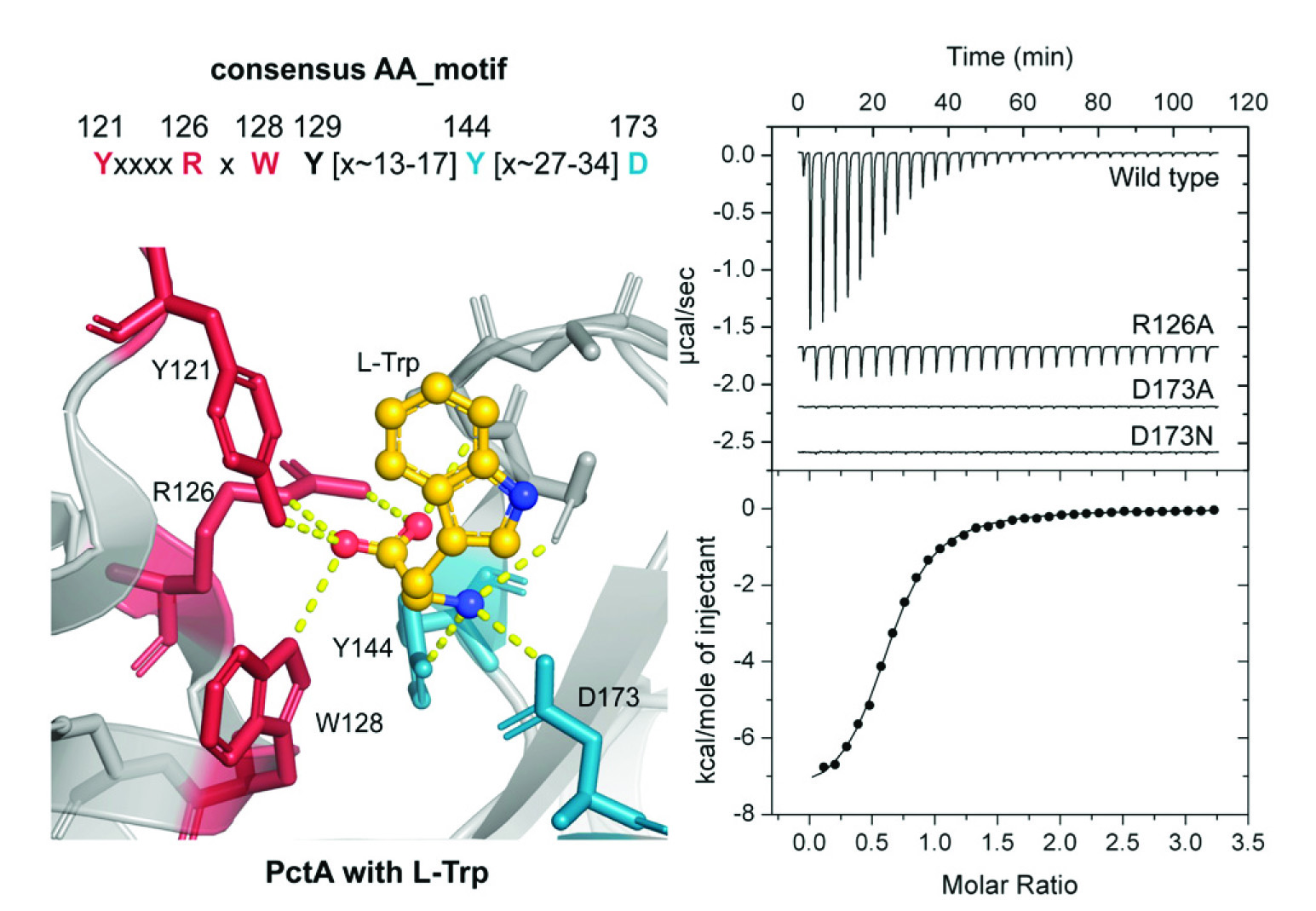
The importance of amino acid sensing for living beings
Gumerov et al. (2022) Proc. Natl. Acad. Sci. USA 119, e2110415119
Organisms throughout the Tree of Life possess the capacity to sense extracellular signals and to adjust a variety of physiological features in response. In this article we discover an extracellular sensing domain, termed dCache_1AA, that recognizes specifically amino acids. This domain is found in archaea, bacteria and different eukaryotes including the human, illustrating the importance of amino acid sensing throughout the Tree of Life. In bacteria, dCache_1AA domains are found in all major families of transmembrane signal transduction receptors including chemoreceptors, sensor histidine kinases, guanylate/adenylate cyclases, c-di-GMP phosphodiesterases or serine/threonine kinases and phosphatases, which underlines its universality. This study has also demonstrated the feasibility of predicting the ligands recognized by sensor domains taking into account the amino acids present in the ligand binding pocket.
Selected publications
Velando, F., Xing, J., Genova, R., Cerna-Vargas, J.P., Vázquez-Santiago, R., Matilla, M.A., Zhulin, I.B., Krell, T. (2025) Chemoreceptor family in plant-associated bacteria responds preferentially to the plant signal molecule glycerol 3-phosphate. Genome Biol. 26:260
Monteagudo-Cascales, E., Gavira, J.A., Xing, J., Velando F., Matilla, M.A., Zhulin, I.B., Krell, T. (2025) Bacterial sensor evolved by decreasing complexity. Proc Natl Acad Sci USA 122(5):e2409881122.
Monteagudo-Cascales, E., Gumerov, V.M., Fernández, M., Matilla, M.A., Gavira, J.A., Zhulin, I.B., Krell, T. (2024) Ubiquitous purine sensor modulates diverse signal transduction pathways in bacteria. Nat Commun. 2024 Jul 12;15(1):5867.
Cerna-Vargas, J.P., Gumerov, V.M., Krell, T., Zhulin, I.B. (2023) Amine-recognizing domain in diverse receptors from bacteria and archaea evolved from the universal amino acid sensor. Proc Natl Acad Sci USA. 120:e2305837120. (Comment article: Dlakić M. 2023. Proc Natl Acad Sci USA 120:e2316830120)
Xu, W. Cerna-Vargas, J.P., Tajuelo, A., Lozano-Montoya, A., Kivoloka, M., Krink, N., Monteagudo-Cascales, E., Matilla, M.A., Krell. T, Sourjik V. (2023) Systematic mapping of chemoreceptor specificities for Pseudomonas aeruginosa. mBio 14:e0209923.
Gavira, J.A., Rico-Jiménez, M., Ortega, Á., Petukhova, N.V., Bug, D.S., Castellví, A., Porozov, Y.B., Zhulin, I.B., Krell, T., Matilla, M.A. (2023) Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria. mBio 14:e0336322.
Monteagudo-Cascales, E., Martín-Mora, D., Xu, W., Sourjik, V., Matilla, M.A. Ortega, A., Krell, T. (2022) The pH robustness of bacterial sensing. mBio e0165022.
Feng, H., Lv, Y., Krell, T., Fu, R., Liu, Y., Xu, Z., Du, W., Shen, Q., Zhang, N. Zhang, R. (2022) Signal binding at both modules of its dCache sensor domain enables the McpA chemoreceptor of Bacillus velezensis to sense different ligands. Proc. Natl. Acad. Sci USA 119:e2201747119
Gumerov, V.M., Andrianova, E.P., Matilla, M.A., Page, K.M., Monteagudo-Cascales, E. Dolphin, A.C., Krell, T. Zhulin, I.B. (2022) Amino acid sensor from bacteria to humans. Proc. Natl. Acad. Sci. USA 119: e2110415119.
Matilla, M.A., Velando, F., Tajuelo, A., Martín-Mora, D., Xu, W., Sourjik, V., Gavira, J.A., Krell, T. (2022) Chemotaxis of the human pathogen Pseudomonas aeruginosa to the neurotransmitter acetylcholine. mBio: e0345821
Matilla, M.A., Velando, F., Martín-Mora, D., Monteagudo-Cascales, E., Krell, T. (2022) A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol Rev. 10.1093/femsre/fuab043
Matilla, M.A., Martín-Mora, D., Gavira, J.A., Krell, T. (2021) Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol. Mol. Biol. Rev. 85(1):e00151-20
Gavira JA, Gumerov VM, Rico-Jiménez M, Petukh M, Upadhyay AA, Ortega A, Matilla MA, Zhulin IB, Krell T. (2020) How Bacterial chemoreceptors evolve novel ligand specificities. mBio 11:e03066-19.
Martín-Mora, D., Ortega, A., Matilla, M.A., Martínez-Rodríguez, S., Gavira, J.A., Krell, T. (2019) The molecular mechanism of nitrate chemotaxis via direct ligand binding to the PilJ domain of McpN. mBio 10:e02334-18
Cerna-Vargas, J.P., Santamaría-Hernando, S., Matilla, M.A., Rodríguez-Herva, J.J., Daddaoua, A., Rodríguez-Palenzuela, P., Krell, T. López-Solanilla, E. (2019) Chemoperception of specific amino acids controls phytopathogenicity in P. syringae pv. Tomato. mBio 10:e01868-19.
Corral-Lugo A., Matilla M.A., Martín-Mora D., Silva Jiménez H., Mesa Torres N., Kato J., Hida A., Oku S., Conejero-Muriel M., Gavira J.A., Krell T. (2018). High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. mBio 9:e01894-18
Matilla M.A., Daddaoua A., Chini A., Morel B., Krell T. (2018). An auxin controls bacterial antibiotics production. Nucleic Acids Res. 46:11229-11238 46:11229-11238
Corral-Lugo A, Daddaoua A, Ortega A, Espinosa-Urgel M, Krell T. (2016) Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Science Signaling 9(409):ra1.
García Fontana, C., Corral-Lugo, A., Krell, T. (2014) Specificity of the CheR2 Methyltransferase in Pseudomonas aeruginosa is Directed by C-Terminal Pentapeptides in Chemoreceptors. Science Signaling 7 (320) ra34.
Pineda-Molina, E., Reyes-Darias, J.A., Lacal, J., Ramos, J.L., García-Ruiz, J.M., Gavira, J.A., Krell, T. (2012) Evidence for chemoreceptors with bimodular ligand binding regions harboring two signal-binding sites. Proc. Acad. Natl. Sci. USA. 109, 18926-18931.


